Integrative visualization of stratified heterogeneous data for disease subtype analysis.
Identification and characterization of subtypes in diseases such as cancer can lead to refined therapeutic targeting, which is an important step towards improved patient outcomes. Integrated analysis of different molecular data types can be used to discover subtypes and suggest molecular differences relevant for therapeutic approaches.
Large-scale data analysis systems developed to perform comprehensive analyses, have strong algorithmic capabilities and generate analysis results required for subtype identification and characterization. The output of these pipelines, however, is often not easy to browse and understand and lacks support for drawing conclusions across datasets and datatypes. Also, automatic methods and statistical tests typically require a specific hypothesis to evaluate and are not well suited to open exploration of data, which is critical to generate hypothesis in the first place.
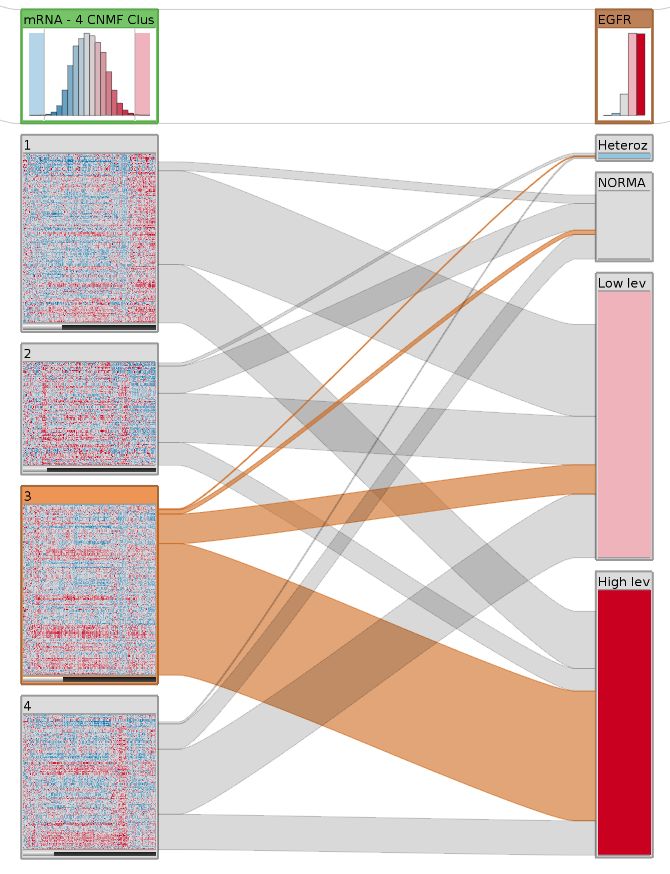
StratomeX addresses these shortcomings by providing an interactive interface that enables analyst to quickly explore complex and heterogeneous datasets. The figure on the right shows a basic StratomeX example. Here, the left column shows a clustered mRNA expression dataset (glioblastoma multiforme). The top block shows the overall distribution, the four blocks below represent groups of patients. Next to it is a column with patients stratified by copy number status. We see that there are many patients that have either a high or at least some additional copies (dark and light-red blocks) of EGFR. The bands between these columns show how many patients overlap between them. The highlighted Block 3 in the left column has a large overlap with the high level and some overlap with the low level amplifications of EGFR, but almost no patients have normal EGFR copy numbers or even deletions, as shown by the very thin bands.
StratomeX supports a rich set of views for different data types that are seamlessly integrated. In addition to the exploration of stratified tabular datasets, as shown in the previous example, StratomeX also visualizes clinical data, which lets analysts reason about the effects of a particular patient stratification on, for example, disease outcomes. Furthermore, StratomeX also integrates pathways that map other datasets onto their genes, which enables analysts to quickly understand the differences in regulation of specific biological processes between patient groups. Both, clinical data and pathways are shown in this picture.
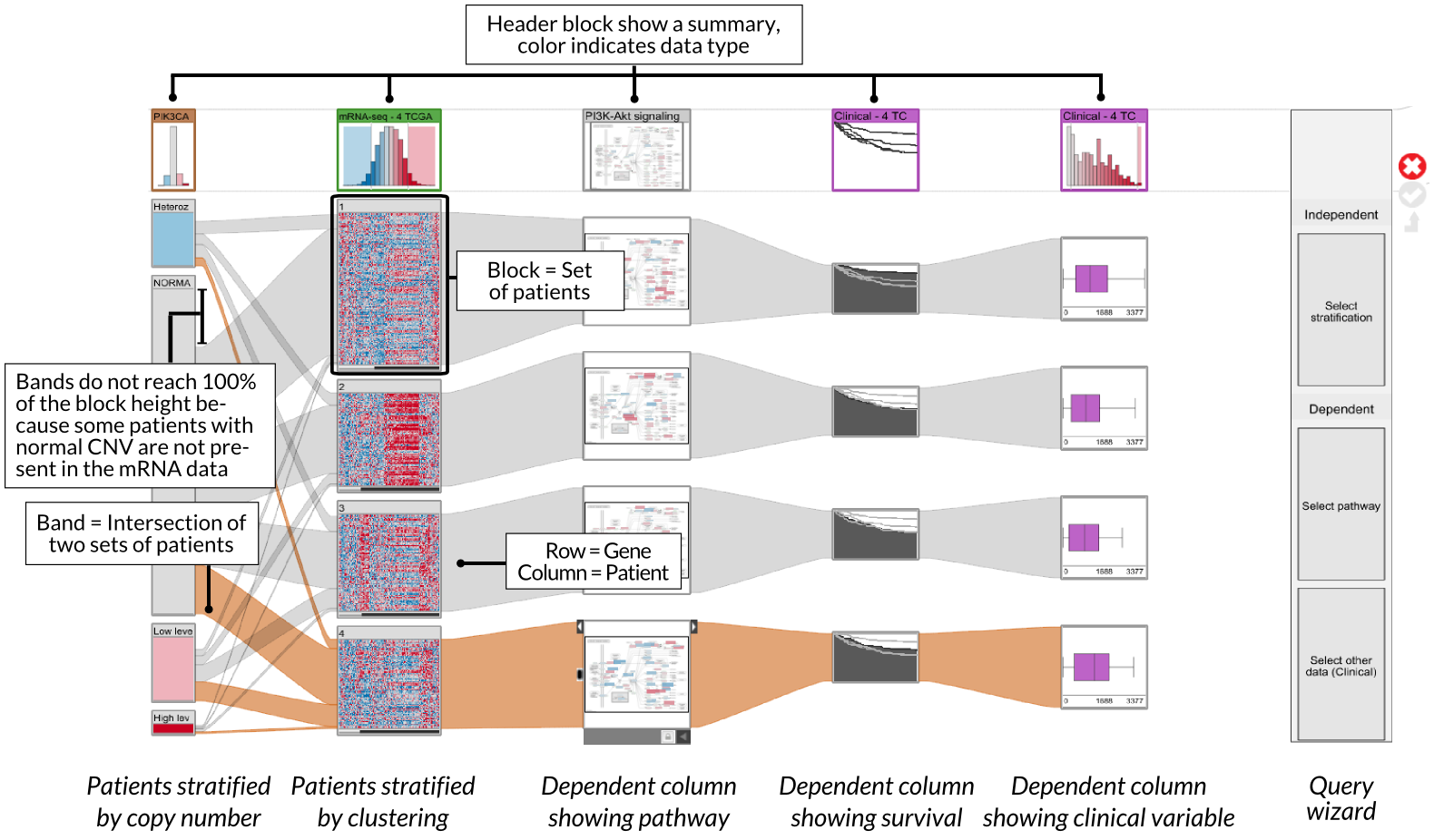
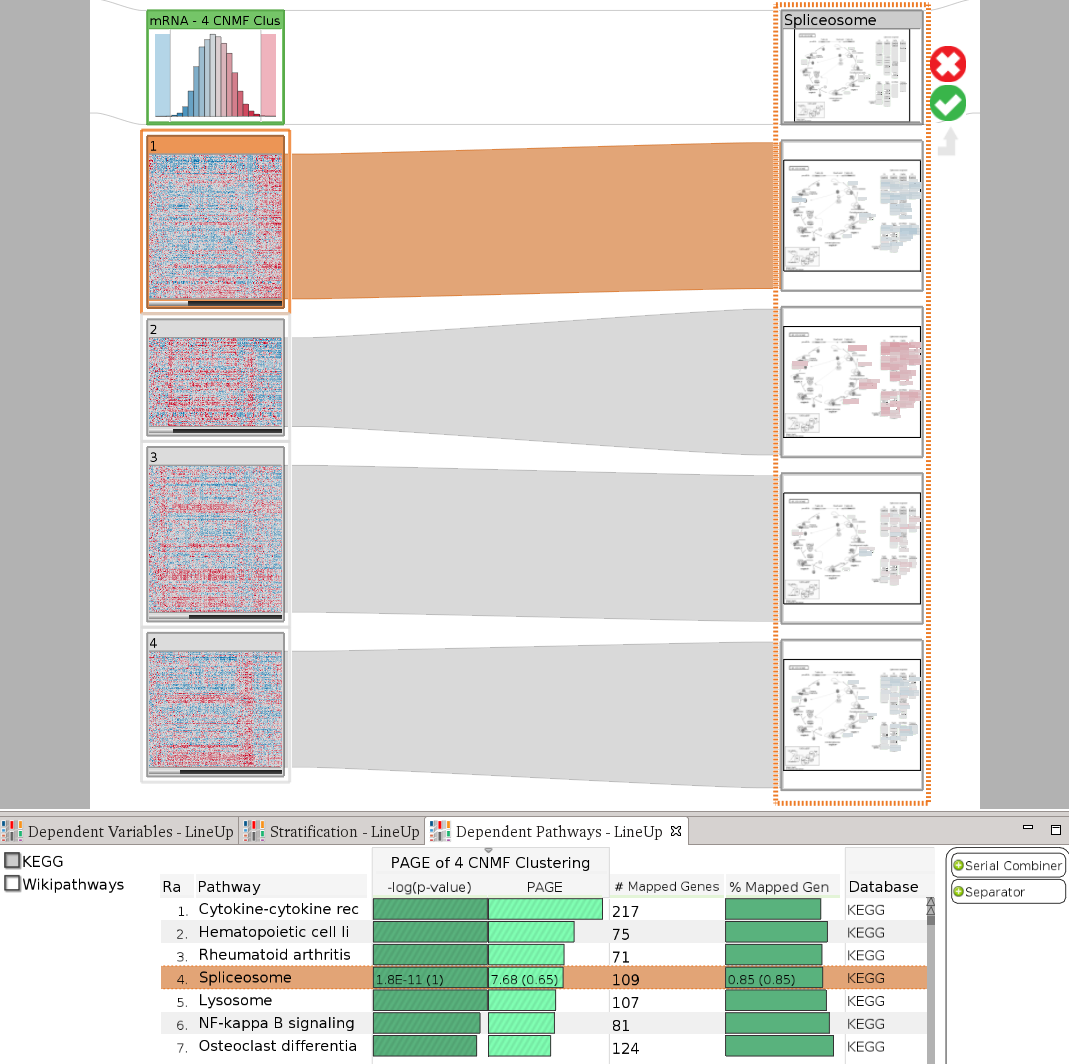
StratomeX also provides an advanced query and data selection interface. The query wizard, shown in the above picture, enables analysts to ask specific questions, such as Which other stratifications are similar to the selected one?, or Is there a pathway that is enriched for this patient group?. A rich set of algorithms calculate rankings of potential matching datasets and stratifications, which are then presented using the LineUp interface. The following example shows a query for a pathway enrichment for an mRNA cluster. LineUp, at the bottom, displays a ranked list of results, of which the fourth is currently selected and displayed.
For more information about StratomeX visit the Help Pages or watch the instructional videos below.
