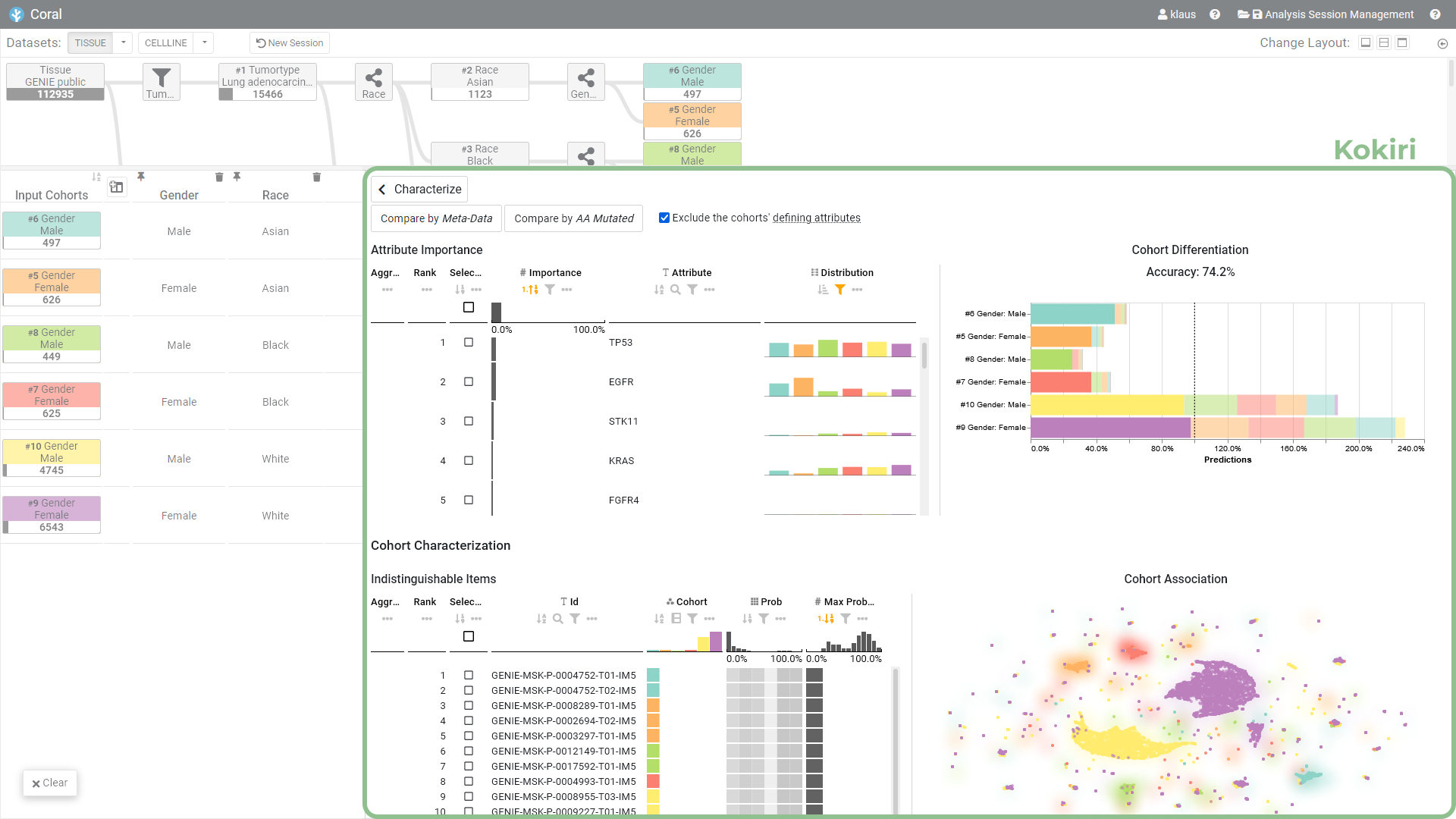
Abstract
We propose an interactive visual analytics approach to characterizing and comparing patient subgroups (i.e., cohorts). Despite having the same disease and similar demographic characteristics, patients respond differently to therapy. One reason for this is the vast number of variables in the genome that influence a patient's outcome. Nevertheless, most existing tools do not offer effective means of identifying the attributes that differ most, or look at them in isolation and thus ignore combinatorial effects. To fill this gap, we present Kokiri, a visual analytics approach that aims to separate cohorts based on user-selected data, ranks attributes by their importance in distinguishing between cohorts, and visualizes cohort overlaps and separability. With our approach, users can additionally characterize the homogeneity and outliers of a cohort. To demonstrate the applicability of our approach, we integrated Kokiri into the Coral cohort analysis tool to compare and characterize lung cancer patient cohorts.
Citation
Klaus
Eckelt,
Patrick
Adelberger,
Markus J. Bauer,
Thomas Zichner,
Marc
Streit
Kokiri: Random-Forest-Based Comparison and Characterization of Cohorts
IEEE VIS Workshop on Visualization in Biomedical AI,
doi:10.1101/2022.08.16.503622, 2022.
BibTeX
@article{2022_kokiri,
title = {Kokiri: Random-Forest-Based Comparison and Characterization of Cohorts},
author = {Klaus Eckelt and Patrick Adelberger and Markus J. Bauer and Thomas Zichner and Marc Streit},
journal = {IEEE VIS Workshop on Visualization in Biomedical AI},
doi = {10.1101/2022.08.16.503622},
url = {https://www.biorxiv.org/content/10.1101/2022.08.16.503622},
year = {2022}
}
Acknowledgements
The authors acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry, and members of the consortium for their commitment to open data. Interpretations are the responsibility of study authors.
This work was supported in part by the Boehringer Ingelheim Regional Center Vienna, the State of Upper Austria, and the Austrian Federal Ministry of Education, Science and Research via the LIT – Linz Institute of Technology (LIT-2019-7-SEE-117), the State of Upper Austria (Human-Interpretable Machine Learning), and the Austrian Science Fund (FWF DFH 23-N).



