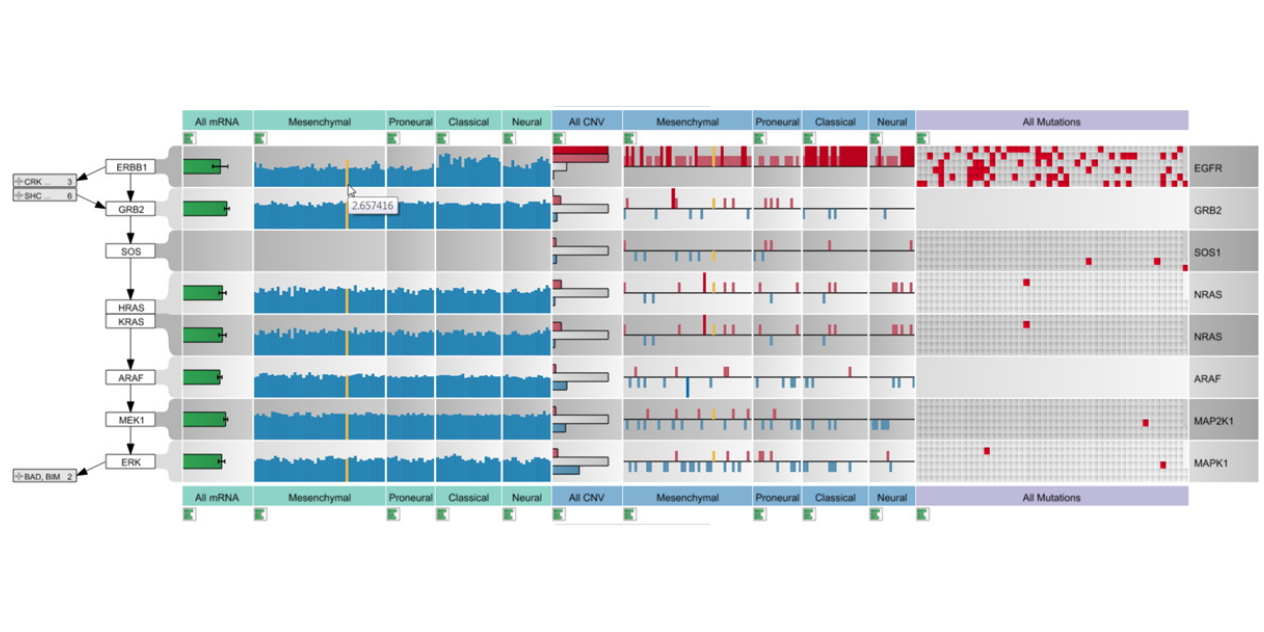
Abstract
Biological pathway maps are highly relevant tools for many tasks in molecular biology. They reduce the complexity of the overall biological network by partitioning it into smaller manageable parts. While this reduction of complexity is their biggest strength, it is, at the same time, their biggest weakness. By removing what is deemed not important for the primary function of the pathway, biologists lose the ability to follow and understand cross-talks between pathways. Considering these cross-talks is, however, critical in many analysis scenarios, such as judging effects of drugs. In this paper we introduce Entourage, a novel visualization technique that provides contextual information lost due to the artificial partitioning of the biological network, but at the same time limits the presented information to what is relevant to the analyst’s task. We use one pathway map as the focus of an analysis and allow a larger set of contextual pathways. For these context pathways we only show the contextual subsets, i.e., the parts of the graph that are relevant to a selection. Entourage suggests related pathways based on similarities and highlights parts of a pathway that are interesting in terms of mapped experimental data. We visualize interdependencies between pathways using stubs of visual links, which we found effective yet not obtrusive. By combining this approach with visualization of experimental data, we can provide domain experts with a highly valuable tool. We demonstrate the utility of Entourage with case studies conducted with a biochemist who researches the effects of drugs on pathways. We show that the technique is well suited to investigate interdependencies between pathways and to analyze, understand, and predict the effect that drugs have on different cell types.
Citation
Christian Partl,
Alexander Lex,
Marc
Streit,
Denis Kalkofen,
Karl Kashofer,
Dieter Schmalstieg
Note: extended journal version of 2012 BioVis article
enRoute: Dynamic Path Extraction from Biological Pathway Maps for Exploring Heterogeneous Experimental Datasets
BMC Bioinformatics,
14(Suppl 19):
S3, doi:10.1186/1471-2105-14-S19-S3, 2013.
BibTeX
@article{2013_bmc_enroute,
title = {enRoute: Dynamic Path Extraction from Biological Pathway Maps for Exploring Heterogeneous Experimental Datasets},
author = {Christian Partl and Alexander Lex and Marc Streit and Denis Kalkofen and Karl Kashofer and Dieter Schmalstieg},
journal = {BMC Bioinformatics},
doi = {10.1186/1471-2105-14-S19-S3},
url = {https://www.biomedcentral.com/1471-2105/14/S19/S3/abstract},
volume = {14},
number = {Suppl 19},
pages = {S3},
month = {nov},
year = {2013}
}

