Abstract
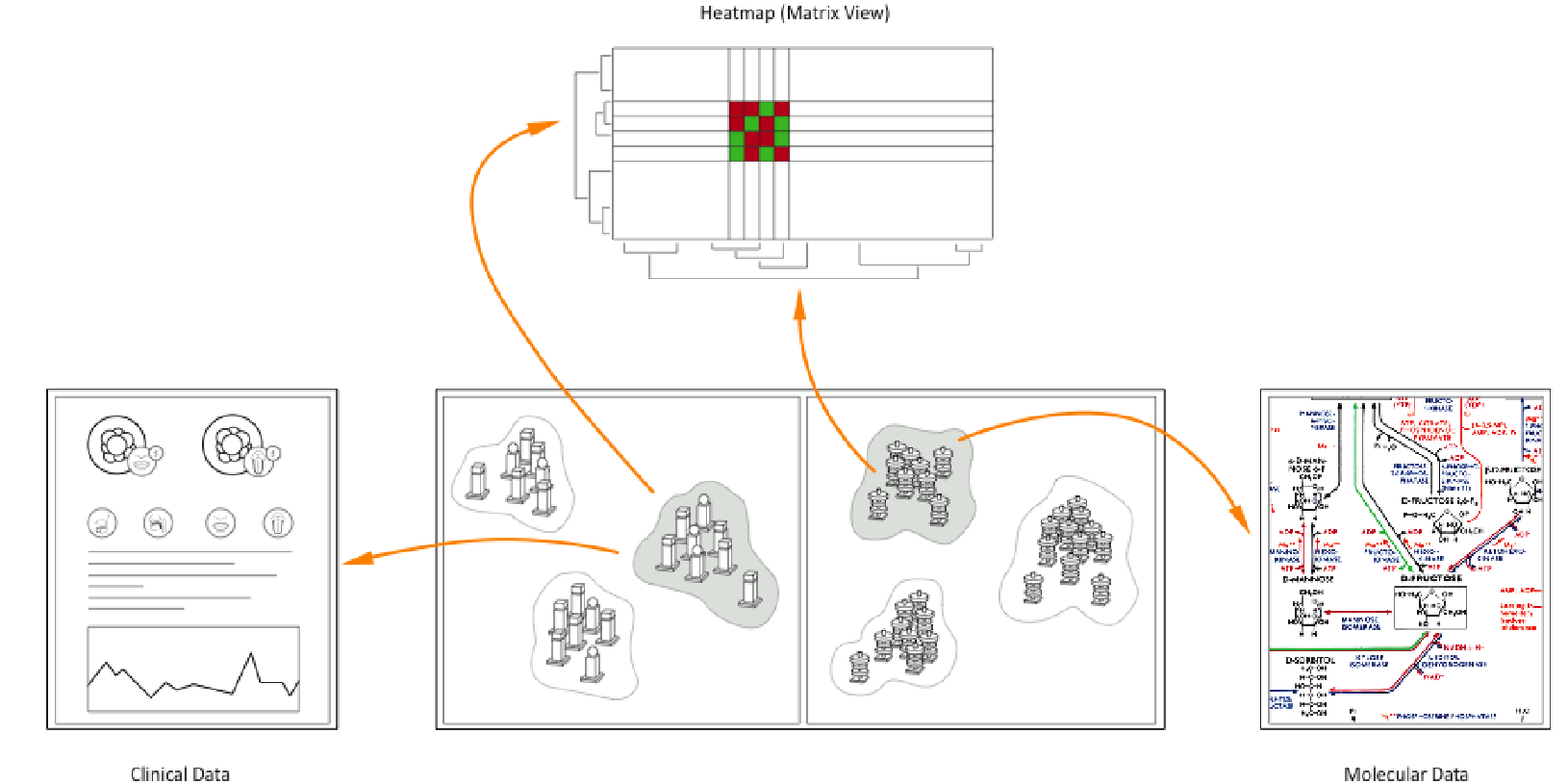
This paper describes an interactive data exploration system for molecular and clinical data in the field of personalized medicine. It addresses the essential but to date unsolved problem of how to identify connections between genetic variants and their corresponding diseases or the response to certain drugs and treatments, respectively. It is therefore necessary to connect genetic with clinical data in order to categorize specific subgroups of patients with certain disease features. The huge amount of data provided by molecular analytical methods (e.g. data on genetic alterations, proteomic or metabolomic data) can only be analyzed by applying statistical methods and bioinformatics. However, even standard methods of statistics and bioinformatics fail when the data is inhomogeneous – as is the case with clinical data – and when data structures are obscured by noise and dominant patterns. The structure of large medical data sets is made visible by using so called object- and attribute-glyphs, which can be arranged in a two dimensional space and synchronized with a set of visualization views.
Citation
Heimo Müller,
Kurt Zatloukal,
Marc
Streit,
Dieter Schmalstieg
Interactive Exploration of Medical Data Sets
Proceedings of the Conference on BioMedical Visualisation,
29--35, doi:10.1109/MediVis.2008.13, 2008.
BibTeX
@article{2008_medivis_medical-glyphs,
title = {Interactive Exploration of Medical Data Sets},
author = {Heimo Müller and Kurt Zatloukal and Marc Streit and Dieter Schmalstieg},
journal = {Proceedings of the Conference on BioMedical Visualisation},
doi = {10.1109/MediVis.2008.13},
pages = {29--35},
year = {2008}
}
Acknowledgements
This work was supported by grants from Styrian Zukunftsfonds, the FIT-IT program of the Austria research funding agency FFG and the Austrian Genome Program (GEN-AU).

